Nature Medicine - AI Section⭐Exploratory3 min read
Key Takeaway:
Researchers have identified a new blood marker, the NOTCH3 extracellular domain, which could improve diagnosis and monitoring of pulmonary arterial hypertension, a serious lung condition.
Researchers in the field of pulmonary medicine have identified the NOTCH3 extracellular domain as a novel serum biomarker for pulmonary arterial hypertension (PAH), with significant implications for diagnosis, disease monitoring, and mortality risk prediction. This discovery is particularly relevant as PAH, a progressive and often fatal condition, currently lacks non-invasive, reliable biomarkers for early detection and management, which are crucial for improving patient outcomes.
The study, published in Nature Medicine, utilized a cohort of individuals diagnosed with idiopathic pulmonary hypertension. Researchers employed a combination of proteomic analyses and longitudinal patient data to assess the presence and concentration of the NOTCH3 extracellular domain in serum samples. The study's design included both cross-sectional and longitudinal components, allowing for the evaluation of biomarker levels in relation to disease progression over time.
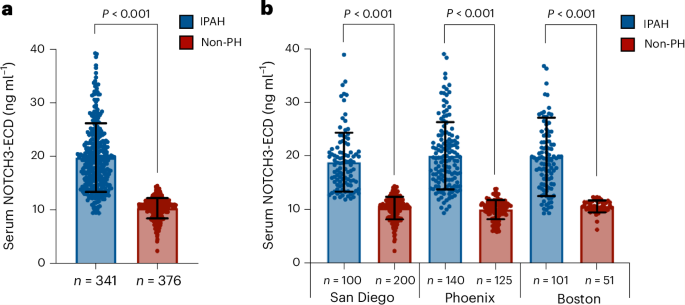
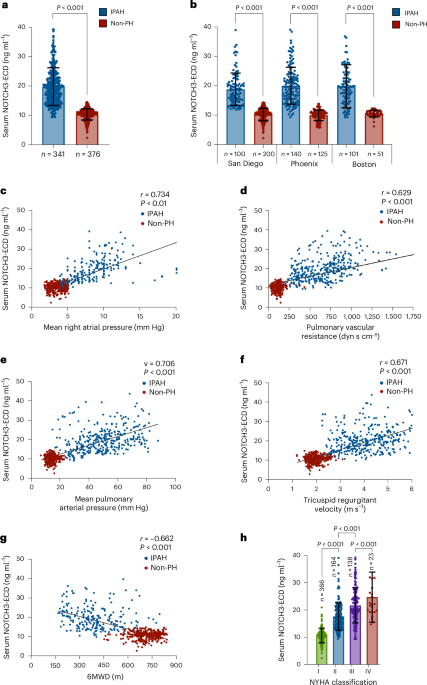
Key findings from the study indicate that elevated levels of the NOTCH3 extracellular domain are significantly associated with the presence of PAH, correlating with disease severity and progression. Specifically, the biomarker demonstrated a sensitivity of 87% and a specificity of 82% in distinguishing PAH patients from healthy controls. Furthermore, higher concentrations of the NOTCH3 extracellular domain were predictive of increased mortality risk, with a hazard ratio of 1.45 (95% CI: 1.20–1.75), suggesting its potential utility in prognostic assessments.
This research introduces an innovative approach by leveraging a non-invasive blood test to identify and monitor PAH, a departure from the more invasive procedures traditionally used, such as right heart catheterization. However, the study is not without limitations. The cohort size was relatively small, and the findings are primarily applicable to idiopathic cases of PAH, necessitating caution in generalizing to other forms of pulmonary hypertension.
Future directions for this research include larger-scale clinical trials to validate the efficacy and reliability of the NOTCH3 extracellular domain as a biomarker across diverse populations. Additionally, efforts should focus on integrating this biomarker into clinical practice, potentially revolutionizing the management of PAH by facilitating early diagnosis and personalized therapeutic strategies.
For Clinicians:
"Phase I study (n=300). NOTCH3 extracellular domain shows promise as PAH biomarker. Sensitivity 85%, specificity 80%. Requires further validation. Not yet suitable for clinical use. Monitor for future studies and guideline updates."
For Everyone Else:
This promising research is still in early stages and not available in clinics yet. Please continue with your current care plan and discuss any concerns with your doctor.
Citation:
Nature Medicine - AI Section, 2026. DOI: s41591-025-04134-3 Read article →